Multivariate Forecasting
This notebook outlines an example of scalecast using multiple series to forecast one another using scikit-learn models.
[1]:
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib
import matplotlib.ticker as ticker
import matplotlib.pyplot as plt
from scalecast.Forecaster import Forecaster
from scalecast.MVForecaster import MVForecaster
from scalecast.multiseries import export_model_summaries
from scalecast import GridGenerator
[2]:
# read data
data = pd.read_csv('avocado.csv',parse_dates=['Date']).sort_values(['Date'])
# sort appropriately (not doing this could cause issues)
data = data.sort_values(['region','type','Date'])
We will be forecasting the organic and conventional avocado sales from California only.
[3]:
data_cali = data.loc[data['region'] == 'California']
data_cali_org = data_cali.loc[data_cali['type'] == 'organic']
data_cali_con = data_cali.loc[data_cali['type'] == 'conventional']
Choose Models and Import Validation Grids
[4]:
# download template validation grids (will not overwrite existing Grids.py file by default)
models = ('mlr','elasticnet','knn','rf','gbt','xgboost','mlp')
GridGenerator.get_example_grids()
GridGenerator.get_mv_grids()
EDA
Plot
[5]:
fig, ax = plt.subplots(figsize=(12,6))
sns.lineplot(
x='Date',
y='Total Volume',
data=data_cali_con,
label='conventional',
ax=ax,
color='black',
legend=False
)
plt.ylabel('Conventional',size=16)
ax2 = ax.twinx()
sns.lineplot(
x='Date',
y='Total Volume',
data=data_cali_org,
label='organic',
ax=ax2,
color='#B2C248',
legend=False
)
ax.figure.legend()
plt.ylabel('Organic',size=16)
ax.yaxis.set_major_formatter(ticker.StrMethodFormatter('{x:,.0f}'))
ax2.yaxis.set_major_formatter(ticker.StrMethodFormatter('{x:,.0f}'))
plt.suptitle('Conventional and Organic Avocado Sale Volumes',size=20)
plt.show()
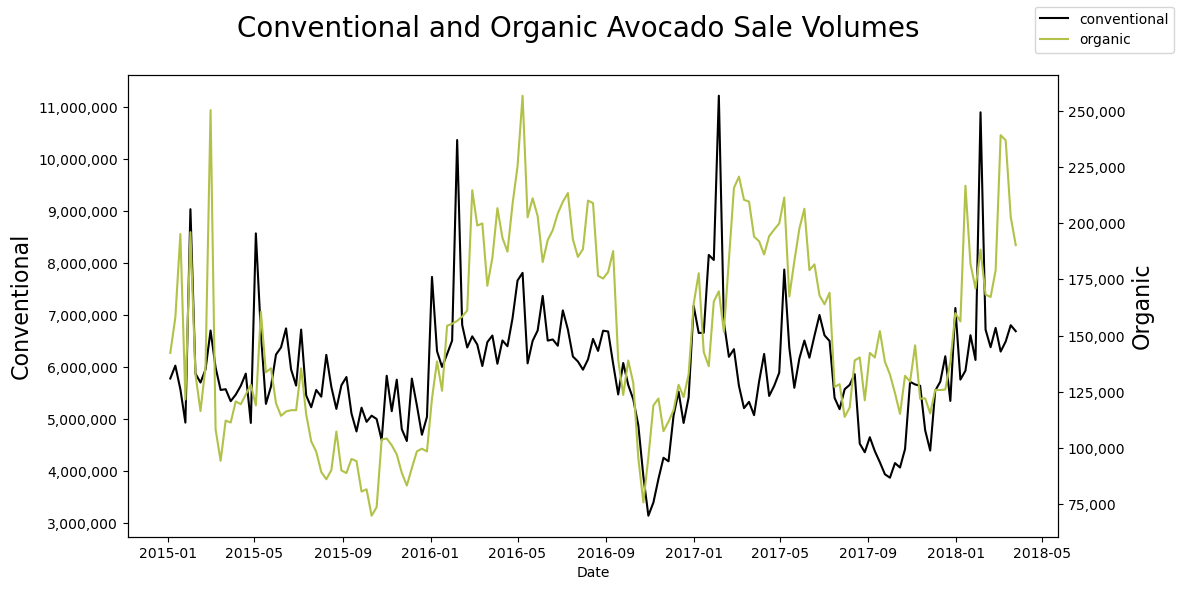
Examine Correlation between the series
[6]:
corr = np.corrcoef(data_cali_org['Total Volume'].values,data_cali_con['Total Volume'].values)[0,1]
print('{:.2%}'.format(corr))
48.02%
Load into Forecaster from scalecast
[7]:
fcon = Forecaster(
y=data_cali_con['Total Volume'],
current_dates = data_cali_con['Date'],
test_length = .2,
future_dates = 52,
validation_length = 4,
metrics = ['rmse','r2'],
cis = True,
)
fcon
[7]:
Forecaster(
DateStartActuals=2015-01-04T00:00:00.000000000
DateEndActuals=2018-03-25T00:00:00.000000000
Freq=W-SUN
N_actuals=169
ForecastLength=52
Xvars=[]
TestLength=33
ValidationMetric=rmse
ForecastsEvaluated=[]
CILevel=0.95
CurrentEstimator=mlr
GridsFile=Grids
)
[8]:
forg = Forecaster(
y=data_cali_org['Total Volume'],
current_dates = data_cali_org['Date'],
test_length = .2,
future_dates = 52,
validation_length = 4,
metrics = ['rmse','r2'],
cis = True,
)
forg
[8]:
Forecaster(
DateStartActuals=2015-01-04T00:00:00.000000000
DateEndActuals=2018-03-25T00:00:00.000000000
Freq=W-SUN
N_actuals=169
ForecastLength=52
Xvars=[]
TestLength=33
ValidationMetric=rmse
ForecastsEvaluated=[]
CILevel=0.95
CurrentEstimator=mlr
GridsFile=Grids
)
ACF and PACF Plots
[9]:
figs, axs = plt.subplots(2, 2,figsize=(16,8))
fcon.plot_acf(
ax=axs[0,0],
title='Conventional ACF',
lags=26,
color='black'
)
fcon.plot_pacf(
ax=axs[0,1],
title='Conventional PACF',
lags=26,
color='black',
method='ywm'
)
forg.plot_acf(
ax=axs[1,0],
title='Organic ACF',
lags=26,
color='#B2C248'
)
forg.plot_pacf(
ax=axs[1,1],
title='Organic PACF',
lags=26,
color='#B2C248',
method='ywm'
)
plt.show()
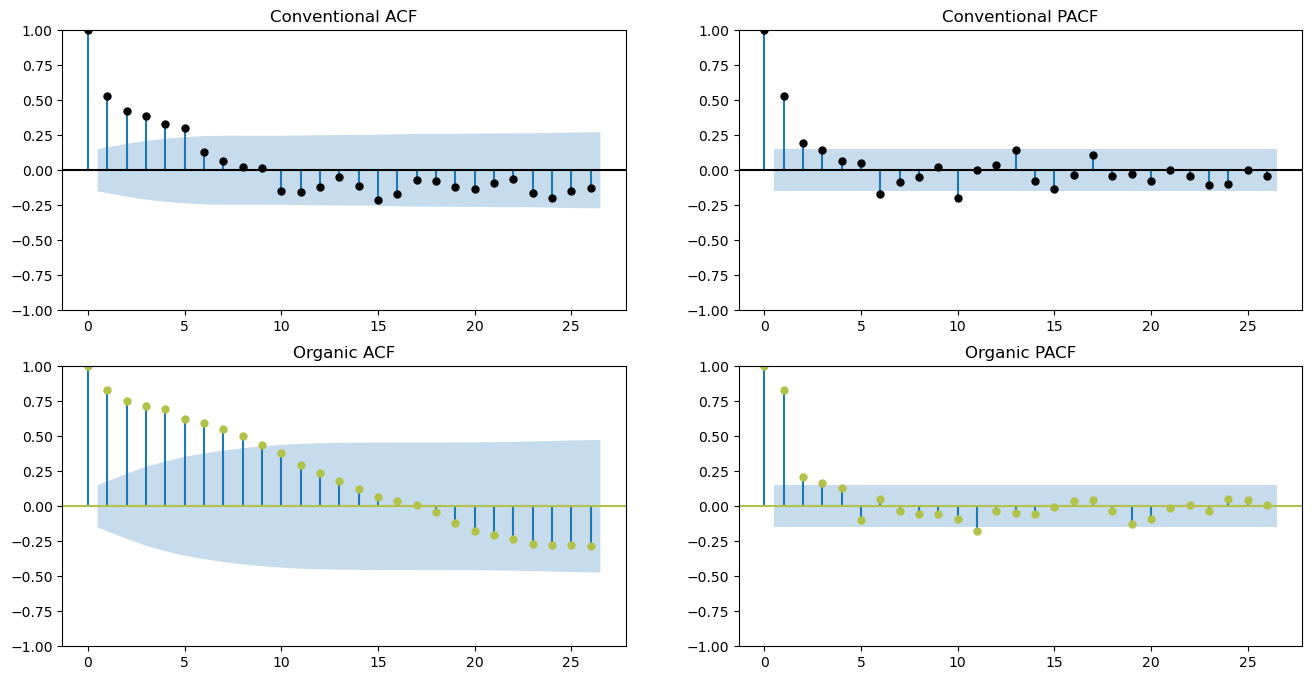
Seasonal Decomposition Plots
[10]:
plt.rc("figure",figsize=(10,6))
fcon.seasonal_decompose(model='mul').plot()
plt.suptitle('Conventional Seasonal Decomp',size=20)
plt.show()
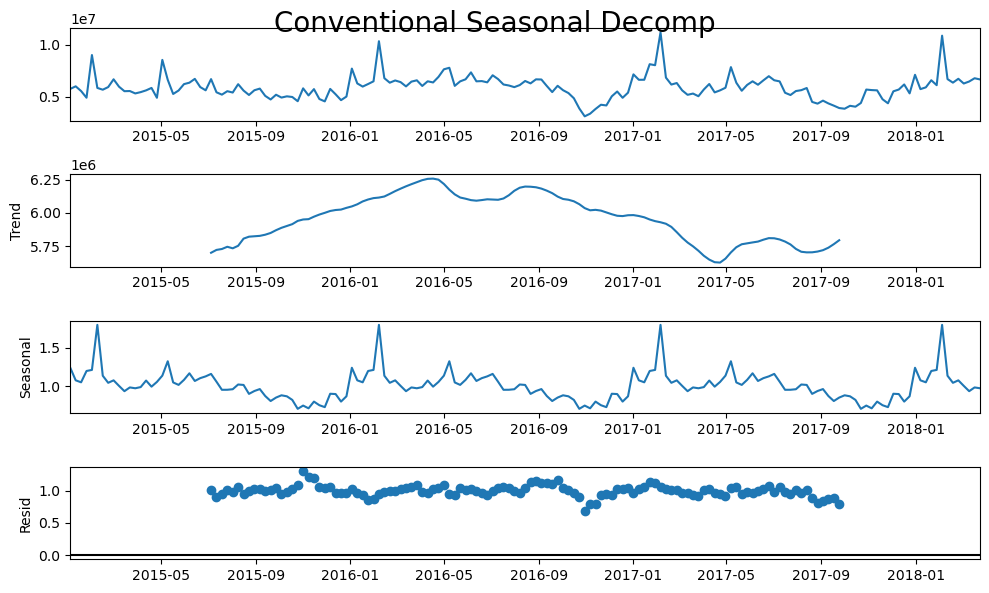
[11]:
forg.seasonal_decompose(model='mul').plot()
plt.suptitle('Organic Seasonal Decomp',size=20)
plt.show()
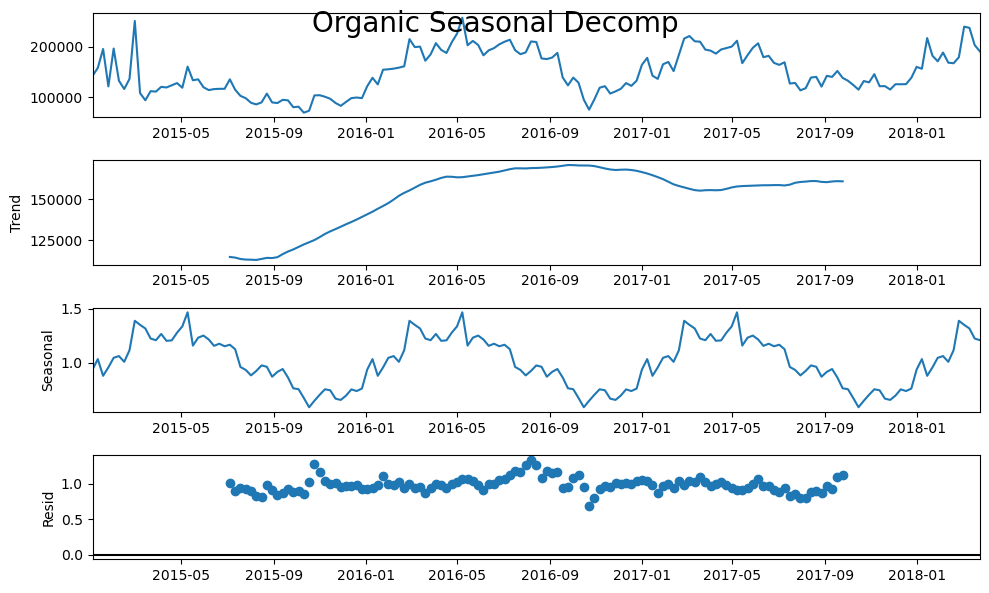
Check Stationarity
[12]:
critical_pval = 0.05
print('-'*100)
print('Conventional Augmented Dickey-Fuller results:')
stat, pval, _, _, _, _ = fcon.adf_test(full_res=True)
print('the test-stat value is: {:.2f}'.format(stat))
print('the p-value is {:.4f}'.format(pval))
print('the series is {}'.format('stationary' if pval < critical_pval else 'not stationary'))
print('-'*100)
print('Organic Augmented Dickey-Fuller results:')
stat, pval, _, _, _, _ = forg.adf_test(full_res=True)
print('the test-stat value is: {:.2f}'.format(stat))
print('the p-value is {:.4f}'.format(pval))
print('the series is {}'.format('stationary' if pval < critical_pval else 'not stationary'))
print('-'*100)
----------------------------------------------------------------------------------------------------
Conventional Augmented Dickey-Fuller results:
the test-stat value is: -3.35
the p-value is 0.0128
the series is stationary
----------------------------------------------------------------------------------------------------
Organic Augmented Dickey-Fuller results:
the test-stat value is: -2.94
the p-value is 0.0404
the series is stationary
----------------------------------------------------------------------------------------------------
Scalecast - Univariate
Load Objects with Xvars:
Find best combination of trend, lags, and seasonality monitoring the validation set (4 observations before the test set)
[13]:
for f in (fcon,forg):
f.auto_Xvar_select(
estimator = 'elasticnet',
monitor = 'ValidationMetricValue',
irr_cycles = [26], # try irregular semi-annual seaosnality
cross_validate=True,
cvkwargs={
'k':3,
'test_length':13,
'space_between_sets':4,
}
)
print(f)
Forecaster(
DateStartActuals=2015-01-04T00:00:00.000000000
DateEndActuals=2018-03-25T00:00:00.000000000
Freq=W-SUN
N_actuals=169
ForecastLength=52
Xvars=['weeksin', 'weekcos', 'monthsin', 'monthcos', 'quartersin', 'quartercos', 'AR1', 'AR2', 'AR3', 'AR4', 'AR5', 'AR6', 'AR7', 'AR8', 'AR9', 'AR10', 'AR11', 'AR12', 'AR13', 'AR14', 'AR15', 'AR16', 'AR17', 'AR18', 'AR19', 'AR20', 'AR21', 'AR22', 'AR23', 'AR24', 'AR25', 'AR26', 'AR27', 'AR28', 'AR29', 'AR30', 'AR31', 'AR32', 'AR33']
TestLength=33
ValidationMetric=rmse
ForecastsEvaluated=[]
CILevel=0.95
CurrentEstimator=mlr
GridsFile=Grids
)
Forecaster(
DateStartActuals=2015-01-04T00:00:00.000000000
DateEndActuals=2018-03-25T00:00:00.000000000
Freq=W-SUN
N_actuals=169
ForecastLength=52
Xvars=['lnt', 'AR1', 'AR2', 'AR3', 'AR4', 'AR5', 'AR6', 'AR7', 'AR8', 'AR9', 'AR10', 'AR11', 'AR12', 'AR13', 'AR14', 'AR15', 'AR16', 'AR17', 'AR18', 'AR19', 'AR20', 'AR21', 'AR22', 'AR23', 'AR24', 'AR25', 'AR26', 'AR27', 'AR28', 'AR29', 'AR30', 'AR31', 'AR32', 'AR33']
TestLength=33
ValidationMetric=rmse
ForecastsEvaluated=[]
CILevel=0.95
CurrentEstimator=mlr
GridsFile=Grids
)
Tune and Forecast with Selected Models
Find optimal hyperparameters using the four out-of-sample observations then test results on test set
Conventional
[14]:
fcon.tune_test_forecast(models,feature_importance=True)
best_model_con_uni = fcon.order_fcsts()[0]
fcon.set_estimator('combo')
fcon.manual_forecast(how='weighted')
2023-04-11 15:32:19.815928: I tensorflow/core/platform/cpu_feature_guard.cc:193] This TensorFlow binary is optimized with oneAPI Deep Neural Network Library (oneDNN) to use the following CPU instructions in performance-critical operations: SSE4.1 SSE4.2
To enable them in other operations, rebuild TensorFlow with the appropriate compiler flags.
[15]:
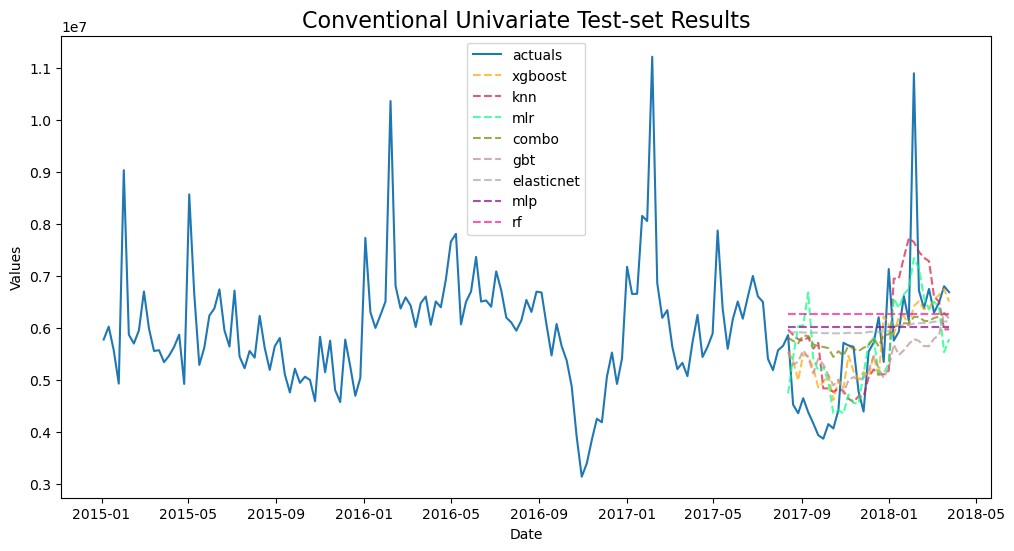
fcon.plot_test_set(order_by='TestSetRMSE')
plt.title('Conventional Univariate Test-set Results',size=16)
plt.show()
Organic
[16]:
forg.tune_test_forecast(models,feature_importance=True)
best_model_org_uni = forg.order_fcsts()[0]
forg.set_estimator('combo')
forg.manual_forecast(how='weighted')
[17]:
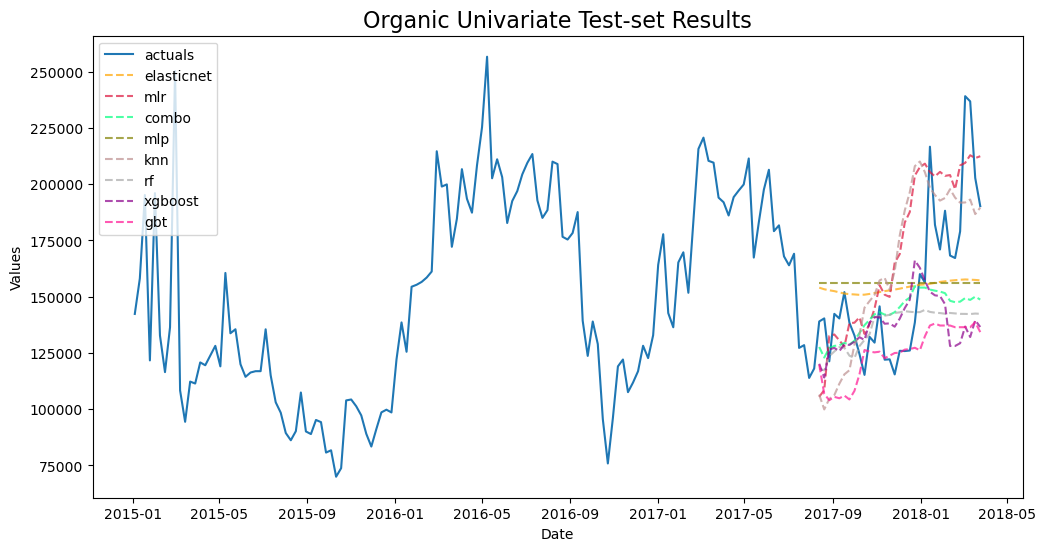
forg.plot_test_set(order_by='TestSetRMSE')
plt.title('Organic Univariate Test-set Results',size=16)
plt.show()
Model Summaries
[22]:
pd.set_option('display.float_format', '{:.4f}'.format)
ms = export_model_summaries({'Conventional':fcon,'Organic':forg},determine_best_by='TestSetRMSE')
ms[
[
'ModelNickname',
'Series',
'TestSetRMSE',
'TestSetR2',
'InSampleRMSE',
'InSampleR2',
'best_model'
]
]
[22]:
| ModelNickname | Series | TestSetRMSE | TestSetR2 | InSampleRMSE | InSampleR2 | best_model | |
|---|---|---|---|---|---|---|---|
| 0 | xgboost | Conventional | 1006995.5599 | 0.4446 | 3.6627 | 1.0000 | True |
| 1 | knn | Conventional | 1131608.0344 | 0.2987 | 817774.1332 | 0.5510 | False |
| 2 | mlr | Conventional | 1149633.3896 | 0.2761 | 758866.6492 | 0.6133 | False |
| 3 | combo | Conventional | 1212096.9382 | 0.1953 | 688989.9601 | 0.6813 | False |
| 4 | gbt | Conventional | 1220551.0383 | 0.1841 | 194073.1900 | 0.9747 | False |
| 5 | elasticnet | Conventional | 1337617.6356 | 0.0201 | 1143622.0298 | 0.1218 | False |
| 6 | mlp | Conventional | 1405812.4388 | -0.0824 | 1220380.8148 | -0.0000 | False |
| 7 | rf | Conventional | 1491918.2868 | -0.2191 | 1223366.9295 | -0.0049 | False |
| 8 | elasticnet | Organic | 31734.3993 | 0.1053 | 37019.4163 | 0.2451 | True |
| 9 | mlr | Organic | 31826.3068 | 0.1001 | 15369.3197 | 0.8699 | False |
| 10 | combo | Organic | 32193.1479 | 0.0793 | 13693.4548 | 0.8967 | False |
| 11 | mlp | Organic | 33604.8672 | -0.0032 | 42607.8305 | -0.0000 | False |
| 12 | knn | Organic | 35461.8762 | -0.1172 | 13801.7064 | 0.8951 | False |
| 13 | rf | Organic | 35785.6551 | -0.1377 | 9688.4987 | 0.9483 | False |
| 14 | xgboost | Organic | 37617.5747 | -0.2571 | 0.6991 | 1.0000 | False |
| 15 | gbt | Organic | 40632.6313 | -0.4667 | 7511.2968 | 0.9689 | False |
Because running so many models can cause overfitting on the test set, we can check the average test error between all models to get another metric of how effective our modeling process is.
[23]:
print('-'*100)
for series in ms['Series'].unique():
print('univariate average test MAPE for {}: {:.4f}'.format(series,ms.loc[ms['Series'] == series,'TestSetRMSE'].mean()))
print('univariate average test R2 for {}: {:.2f}'.format(series,ms.loc[ms['Series'] == series,'TestSetR2'].mean()))
print('-'*100)
----------------------------------------------------------------------------------------------------
univariate average test MAPE for Conventional: 1244529.1652
univariate average test R2 for Conventional: 0.14
----------------------------------------------------------------------------------------------------
univariate average test MAPE for Organic: 34857.0573
univariate average test R2 for Organic: -0.09
----------------------------------------------------------------------------------------------------
These are interesting error metrics, but a lot of models, including XGBoost and GBT appear to be overfitting. Let’s see if we can beat the results with a multivariate approach.
Scalecast - Multivariate
Set MV Parameters
Forecast horizon already set
Xvars already set
Test size must be set: 20%
Validation size must be set: 4 periods
[24]:
mvf = MVForecaster(
fcon,forg,
names=['Conventional','Organic'],
test_length = .2,
valiation_length = 4,
cis = True,
metrics = ['rmse','r2'],
) # init the mvf object
mvf
[24]:
MVForecaster(
DateStartActuals=2015-01-04T00:00:00.000000000
DateEndActuals=2018-03-25T00:00:00.000000000
Freq=W-SUN
N_actuals=169
N_series=2
SeriesNames=['Conventional', 'Organic']
ForecastLength=52
Xvars=['weeksin', 'weekcos', 'monthsin', 'monthcos', 'quartersin', 'quartercos', 'lnt']
TestLength=33
ValidationLength=1
ValidationMetric=rmse
ForecastsEvaluated=[]
CILevel=0.95
CurrentEstimator=mlr
OptimizeOn=mean
GridsFile=MVGrids
)
The lags from each Forecaster object dropped but they will be added into the multivariate models differently.
View Series Correlation
[25]:
mvf.corr()
[25]:
| Conventional | Organic | |
|---|---|---|
| Conventional | 1.0000 | 0.4802 |
| Organic | 0.4802 | 1.0000 |
View Series Correlation with each others’ lags
[26]:
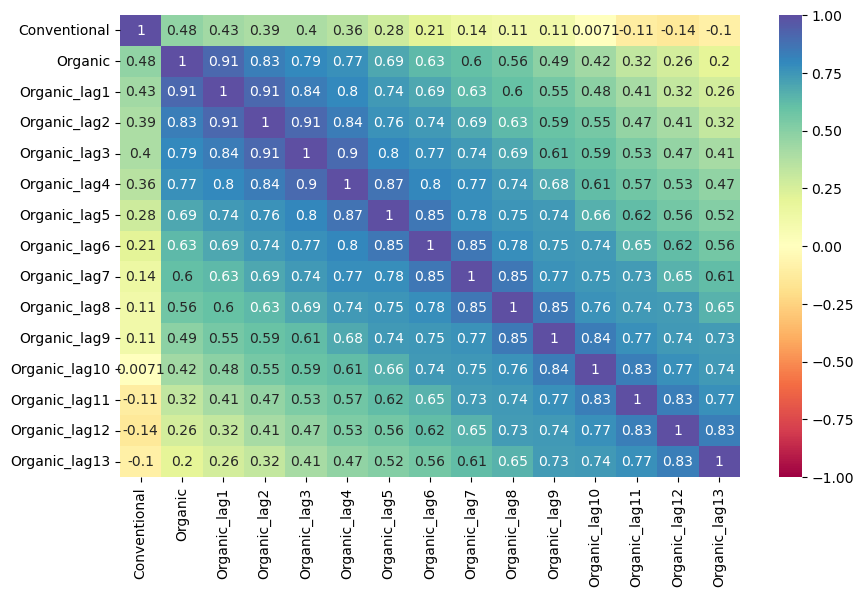
mvf.corr_lags(
y='Conventional',
x='Organic',
lags=13,
disp='heatmap',
annot=True,
vmin=-1,
vmax=1,
cmap = 'Spectral',
)
plt.show()
[27]:
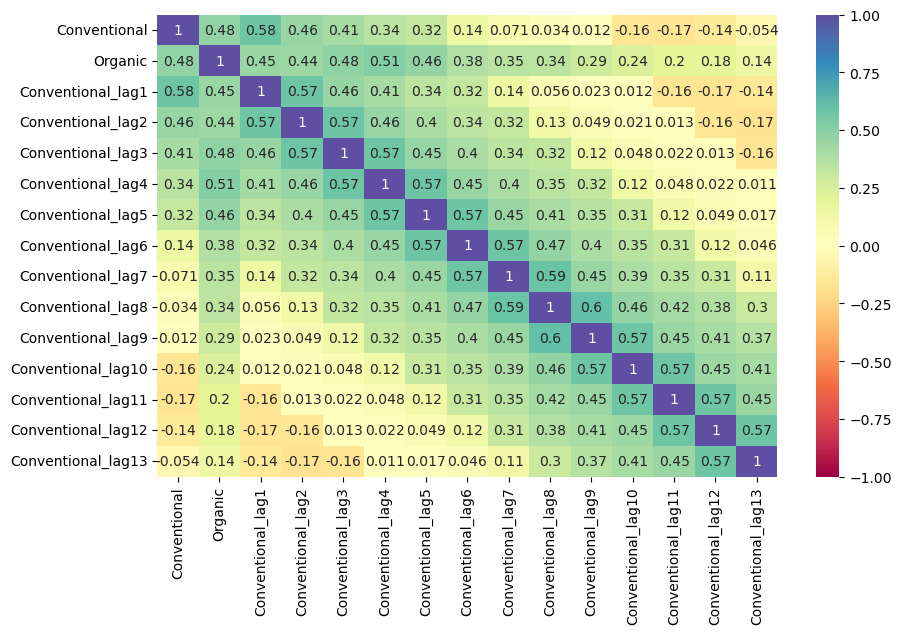
mvf.corr_lags(
y='Organic',
x='Conventional',
lags=13,
disp='heatmap',
annot=True,
vmin=-1,
vmax=1,
cmap = 'Spectral',
)
plt.show()
Set Optimize On
If predicting one series is more important than predicting the other, you can use this code to let the code know to favor one over the other
By default, it uses the mean error metrics between all series
[28]:
# how to optimize on one series
mvf.set_optimize_on('Organic')
# how to optimize using a weighted avarage
mvf.add_optimizer_func(lambda x: x[0]*.25 + x[1]*.75,'weighted')
mvf.set_optimize_on('weighted')
# how to optimize on the average of both/all series (default)
mvf.set_optimize_on('mean')
Tune and Test with Selected Models
[29]:
mvf.tune_test_forecast(models)
mvf.set_best_model(determine_best_by='TestSetRMSE')
[30]:
# not plotting both series at the same time because they have significantly different scales
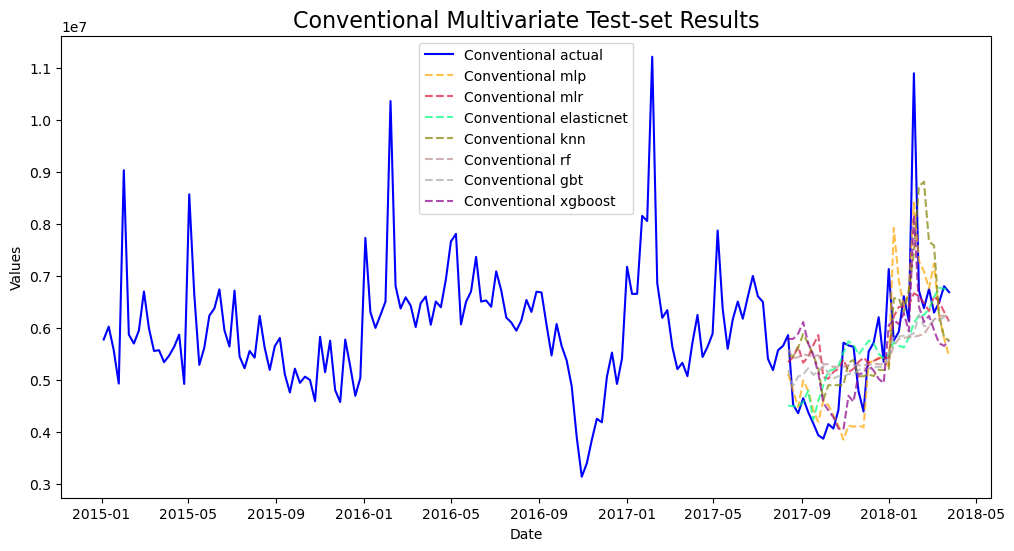
mvf.plot_test_set(series='Conventional',put_best_on_top=True)
plt.title('Conventional Multivariate Test-set Results',size=16)
plt.show()
[31]:
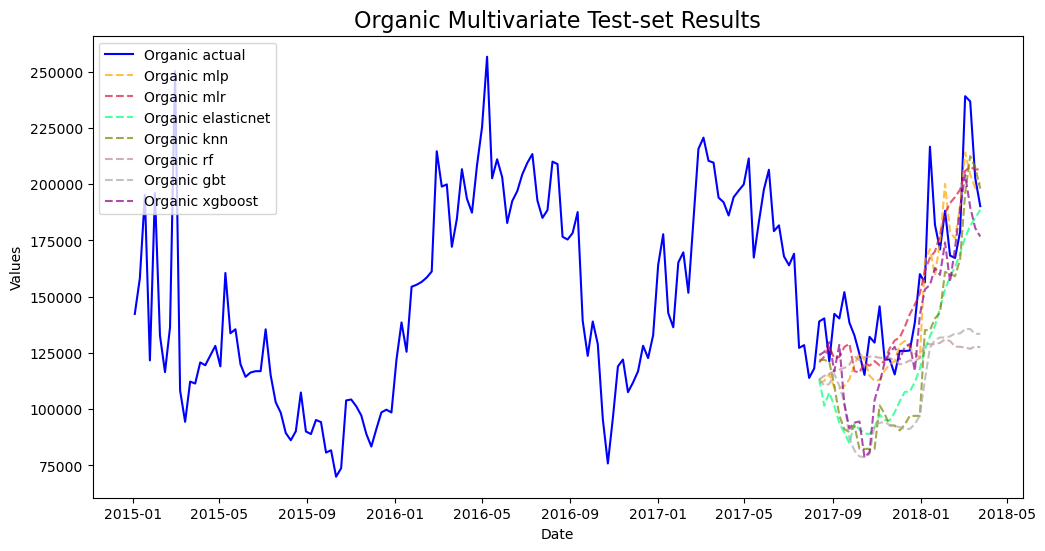
mvf.plot_test_set(series='Organic',put_best_on_top=True)
plt.title('Organic Multivariate Test-set Results',size=16)
plt.show()
Export Model Summaries
[32]:
pd.options.display.max_colwidth = 100
results = mvf.export('model_summaries')
results[
[
'ModelNickname',
'Series',
'HyperParams',
'TestSetRMSE',
'TestSetR2',
'InSampleRMSE',
'InSampleR2',
'Lags'
]
]
[32]:
| ModelNickname | Series | HyperParams | TestSetRMSE | TestSetR2 | InSampleRMSE | InSampleR2 | Lags | |
|---|---|---|---|---|---|---|---|---|
| 0 | mlp | Conventional | {'activation': 'relu', 'hidden_layer_sizes': (25, 25), 'solver': 'lbfgs', 'normalizer': 'minmax'... | 976640.7441 | 0.4776 | 726613.9497 | 0.6134 | 3 |
| 1 | mlr | Conventional | {'normalizer': None} | 1090383.4159 | 0.3488 | 889221.9216 | 0.4209 | 3 |
| 2 | elasticnet | Conventional | {'alpha': 0.9, 'l1_ratio': 0.75, 'normalizer': None} | 1053128.2384 | 0.3926 | 752345.3345 | 0.5889 | 13 |
| 3 | knn | Conventional | {'n_neighbors': 4, 'weights': 'uniform'} | 1178477.0280 | 0.2394 | 663362.5001 | 0.6804 | 13 |
| 4 | rf | Conventional | {'max_depth': 5, 'n_estimators': 100} | 1202344.6297 | 0.2082 | 343671.0745 | 0.9142 | 13 |
| 5 | gbt | Conventional | {'max_depth': 2, 'max_features': None} | 1124594.8764 | 0.3073 | 259436.1165 | 0.9511 | 13 |
| 6 | xgboost | Conventional | {'max_depth': 2} | 974696.0984 | 0.4797 | 233374.4324 | 0.9597 | 1 |
| 7 | mlp | Organic | {'activation': 'relu', 'hidden_layer_sizes': (25, 25), 'solver': 'lbfgs', 'normalizer': 'minmax'... | 20920.8153 | 0.6112 | 16252.8787 | 0.8544 | 3 |
| 8 | mlr | Organic | {'normalizer': None} | 17923.9250 | 0.7146 | 20380.0266 | 0.7710 | 3 |
| 9 | elasticnet | Organic | {'alpha': 0.9, 'l1_ratio': 0.75, 'normalizer': None} | 37629.9902 | -0.2580 | 14968.7668 | 0.8746 | 13 |
| 10 | knn | Organic | {'n_neighbors': 4, 'weights': 'uniform'} | 36796.6649 | -0.2029 | 12551.0264 | 0.9118 | 13 |
| 11 | rf | Organic | {'max_depth': 5, 'n_estimators': 100} | 43547.8368 | -0.6848 | 8941.5674 | 0.9552 | 13 |
| 12 | gbt | Organic | {'max_depth': 2, 'max_features': None} | 50265.9563 | -1.2447 | 7325.5184 | 0.9700 | 13 |
| 13 | xgboost | Organic | {'max_depth': 2} | 26986.2883 | 0.3530 | 6488.8763 | 0.9767 | 1 |
Import a Foreign Sklearn Estimator for Ensemble Modeling
[33]:
from sklearn.ensemble import StackingRegressor
from sklearn.linear_model import LinearRegression
from sklearn.linear_model import ElasticNet
from sklearn.neural_network import MLPRegressor
from sklearn.neighbors import KNeighborsRegressor
from xgboost import XGBRegressor
estimators = [
(
'mlr',
LinearRegression()
),
(
'elasticnet',
ElasticNet(
**{
k:v for k,v in (
results.loc[
results['ModelNickname'] == 'elasticnet',
'HyperParams',
].values[0]
).items() if k != 'normalizer'
}
)
),
(
'mlp',
MLPRegressor(
**{
k:v for k,v in (
results.loc[
results['ModelNickname'] == 'mlp',
'HyperParams',
].values[0]
).items() if k != 'normalizer'
}
)
)
]
final_estimator = KNeighborsRegressor(
**{
k:v for k,v in (
results.loc[
results['ModelNickname'] == 'knn',
'HyperParams',
].values[0]
).items() if k != 'normalizer'
}
)
[34]:
mvf.add_sklearn_estimator(StackingRegressor,'stacking')
mvf.set_estimator('stacking')
mvf.manual_forecast(estimators=estimators,final_estimator=final_estimator,lags=13)
[35]:
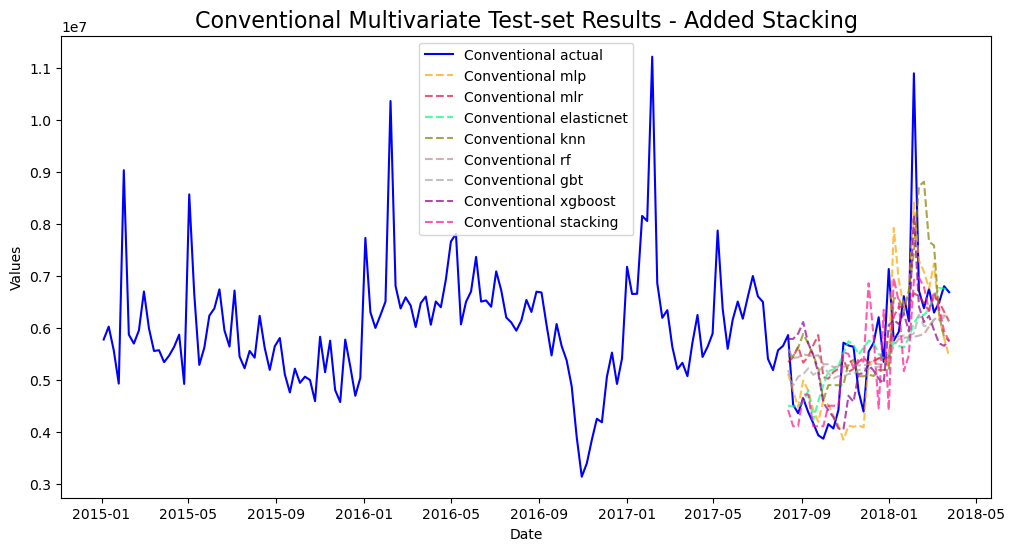
mvf.plot_test_set(series='Conventional',put_best_on_top=True)
plt.title('Conventional Multivariate Test-set Results - Added Stacking',size=16)
plt.show()
[36]:
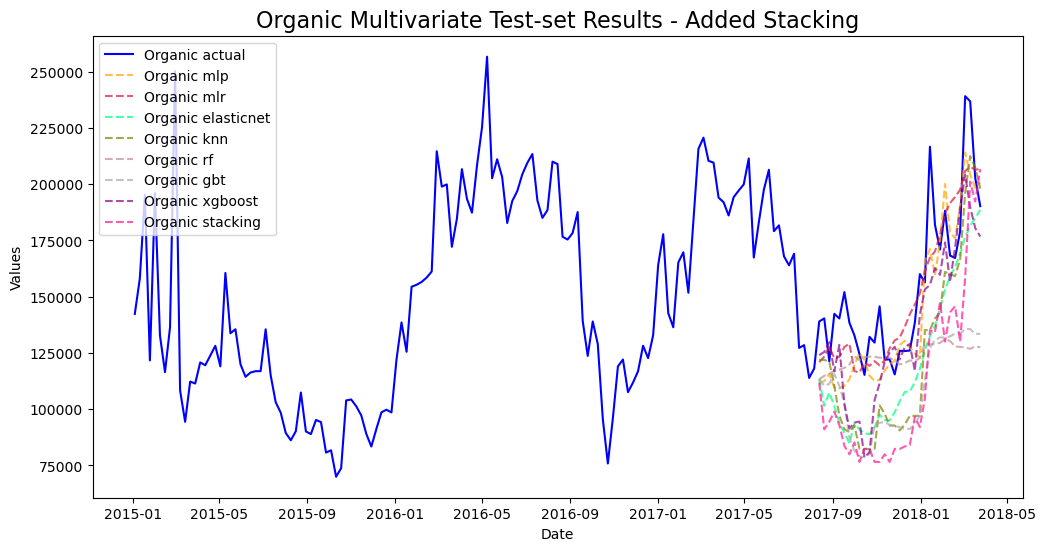
mvf.plot_test_set(series='Organic',put_best_on_top=True)
plt.title('Organic Multivariate Test-set Results - Added Stacking',size=16)
plt.show()
[37]:
mvf.set_best_model(determine_best_by='TestSetRMSE')
results2 = mvf.export('model_summaries')
results2 = results2[
[
'ModelNickname',
'Series',
'TestSetRMSE',
'TestSetR2',
'InSampleRMSE',
'InSampleR2',
'Lags',
'best_model'
]
]
results2
[37]:
| ModelNickname | Series | TestSetRMSE | TestSetR2 | InSampleRMSE | InSampleR2 | Lags | best_model | |
|---|---|---|---|---|---|---|---|---|
| 0 | mlp | Conventional | 976640.7441 | 0.4776 | 726613.9497 | 0.6134 | 3 | True |
| 1 | mlr | Conventional | 1090383.4159 | 0.3488 | 889221.9216 | 0.4209 | 3 | False |
| 2 | elasticnet | Conventional | 1053128.2384 | 0.3926 | 752345.3345 | 0.5889 | 13 | False |
| 3 | knn | Conventional | 1178477.0280 | 0.2394 | 663362.5001 | 0.6804 | 13 | False |
| 4 | rf | Conventional | 1202344.6297 | 0.2082 | 343671.0745 | 0.9142 | 13 | False |
| 5 | gbt | Conventional | 1124594.8764 | 0.3073 | 259436.1165 | 0.9511 | 13 | False |
| 6 | xgboost | Conventional | 974696.0984 | 0.4797 | 233374.4324 | 0.9597 | 1 | False |
| 7 | stacking | Conventional | 1098442.4396 | 0.3392 | 838807.3770 | 0.4890 | 13 | False |
| 8 | mlp | Organic | 20920.8153 | 0.6112 | 16252.8787 | 0.8544 | 3 | True |
| 9 | mlr | Organic | 17923.9250 | 0.7146 | 20380.0266 | 0.7710 | 3 | False |
| 10 | elasticnet | Organic | 37629.9902 | -0.2580 | 14968.7668 | 0.8746 | 13 | False |
| 11 | knn | Organic | 36796.6649 | -0.2029 | 12551.0264 | 0.9118 | 13 | False |
| 12 | rf | Organic | 43547.8368 | -0.6848 | 8941.5674 | 0.9552 | 13 | False |
| 13 | gbt | Organic | 50265.9563 | -1.2447 | 7325.5184 | 0.9700 | 13 | False |
| 14 | xgboost | Organic | 26986.2883 | 0.3530 | 6488.8763 | 0.9767 | 1 | False |
| 15 | stacking | Organic | 47928.2496 | -1.0407 | 17511.0377 | 0.8284 | 13 | False |
[38]:
print('-'*100)
for series in results2['Series'].unique():
print('multivariate average test MAPE for {}: {:.4f}'.format(series,results2.loc[results2['Series'] == series,'TestSetRMSE'].mean()))
print('multivariate average test R2 for {}: {:.2f}'.format(series,results2.loc[results2['Series'] == series,'TestSetR2'].mean()))
print('-'*100)
----------------------------------------------------------------------------------------------------
multivariate average test MAPE for Conventional: 1087338.4338
multivariate average test R2 for Conventional: 0.35
----------------------------------------------------------------------------------------------------
multivariate average test MAPE for Organic: 35249.9658
multivariate average test R2 for Organic: -0.22
----------------------------------------------------------------------------------------------------
Plot Final Forecasts
[39]:
best_model_con = (
results2.loc[results2['Series'] == 'Conventional']
.sort_values('TestSetR2',ascending=False)
.iloc[0,0]
)
[40]:
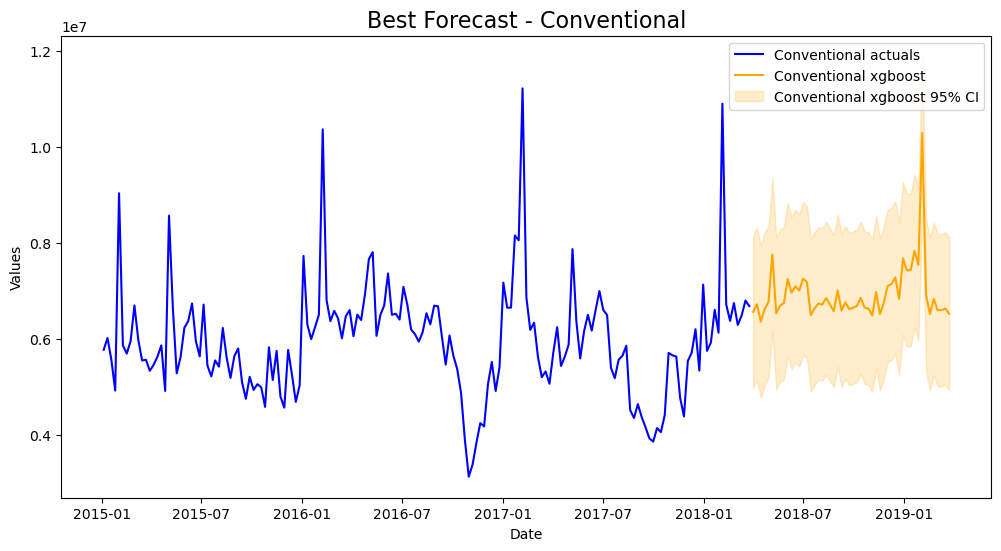
mvf.plot(series='Conventional',models=best_model_con,ci=True)
plt.title('Best Forecast - Conventional',size=16)
plt.show()
[41]:
best_model_org = (
results2.loc[results2['Series'] == 'Organic']
.sort_values('TestSetR2',ascending=False)
.iloc[0,0]
)
[42]:
mvf.plot(series='Organic',models=best_model_org,ci=True)
plt.title('Best Forecast - Organic',size=16)
plt.show()
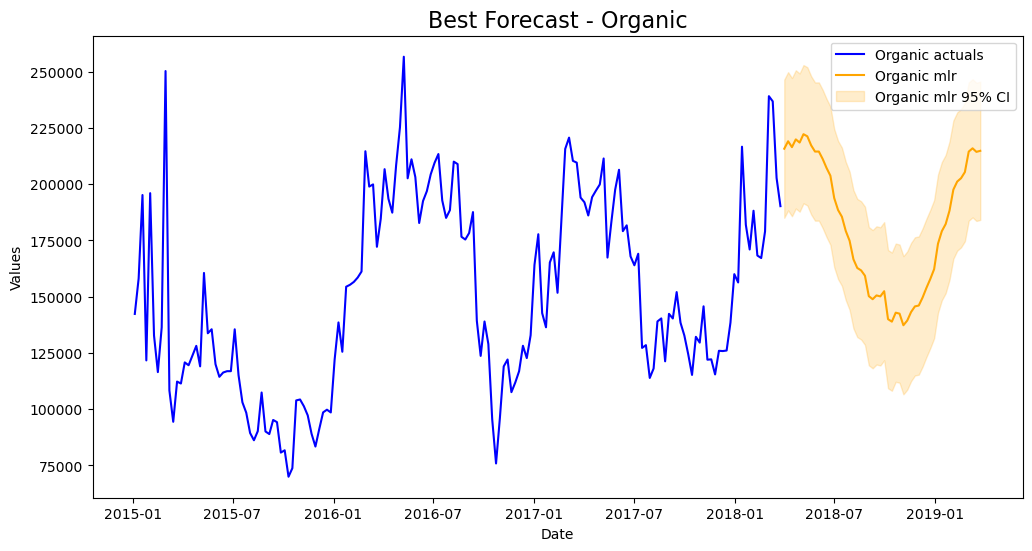
All-in-all, better results on the test-set were obtained by using the multivariate approach.
Multivariate Backtest
Just like in univariate modeling, we can evaluate the results of this modeling approach using backtesting.
[46]:
from scalecast.Pipeline import Transformer, Reverter, MVPipeline
from scalecast.util import backtest_metrics
def mvforecaster(mvf):
mvf.add_seasonal_regressors('month','quarter',raw=False,sincos=True)
mvf.tune_test_forecast(
models=[best_model_con,best_model_org],
)
pipeline = MVPipeline(
steps = [('Forecast',mvforecaster)],
validation_length = 4,
)
[47]:
backtest_results = pipeline.backtest(
fcon,
forg,
cis=False,
test_length = 0,
fcst_length = 52,
jump_back = 4,
n_iter = 3,
)
[48]:
backtest_metrics(
backtest_results,
mets=['rmse','r2'],
names=['Conventional','Organic'],
)
[48]:
| Iter0 | Iter1 | Iter2 | Average | |||
|---|---|---|---|---|---|---|
| Series | Model | Metric | ||||
| Conventional | xgboost | rmse | 1147549.7983 | 1406113.2605 | 1291947.8525 | 1281870.3037 |
| r2 | 0.0323 | -0.4901 | -0.2085 | -0.2221 | ||
| mlr | rmse | 1182695.7635 | 1091062.5392 | 1004623.8802 | 1092794.0610 | |
| r2 | -0.0278 | 0.1028 | 0.2693 | 0.1148 | ||
| Organic | xgboost | rmse | 38107.4049 | 48419.8474 | 50356.9359 | 45628.0627 |
| r2 | -0.3062 | -1.3092 | -1.3834 | -0.9996 | ||
| mlr | rmse | 34053.1727 | 29246.3029 | 21894.3284 | 28397.9347 | |
| r2 | -0.0430 | 0.1575 | 0.5495 | 0.2213 |
[ ]: